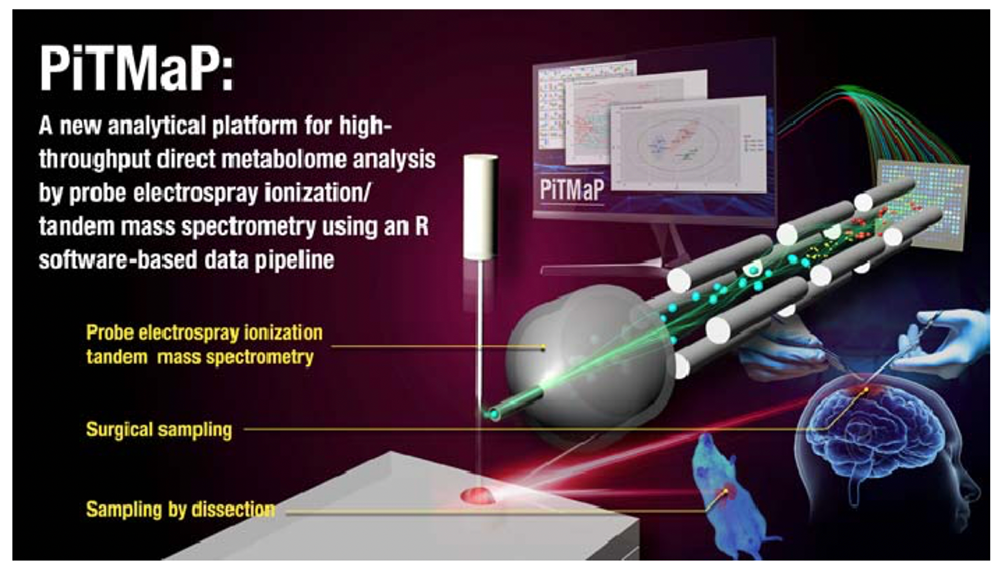
Development of a new analytical platform for high‐throughput direct metabolome analysis by probe electrospray ionization/tandem mass spectrometry using an R software‐based data pipeline
Summary of AIST Press Release May 25, 2020
>>Japanese
The research group of Kei Zaitsu (Nagoya University), Seiichiro Eguchi (Tokyo Women’s Medical University) and Akira Iguchi (Research Institute of Geology and Geoinformation, AIST) has succeeded in developing a new analytical platform for high‐throughput direct metabolome analysis by probe electrospray ionization/tandem mass spectrometry using an R software‐based data pipeline, called PiTMaP (Fig. 1).
Fig. 1. Schematic view of PiTMaP.
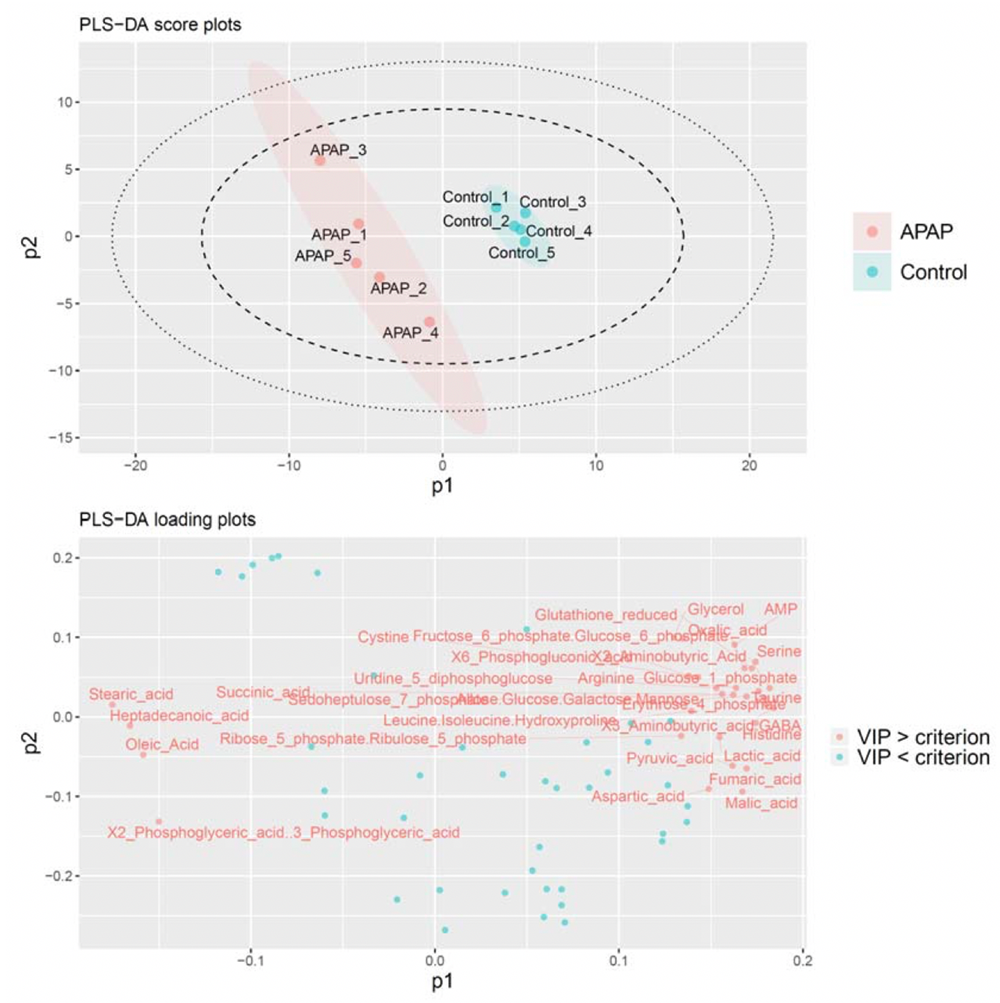
PiTMaP can directly monitor 72 metabolites in tissue samples within 2.4 min per sample without tedious sample preparation. In addition, development of an R software-based data pipeline has enabled multiple tasks to be completed within ca. 1 min: i) automatic box‐and‐whisker plotting for all metabolites, ii) multivariate analyses such as principal component analysis (PCA) and projection to latent structures‐discriminant analysis (PLS‐DA), iii) scoring and loading plots of PCA and PLS‐DA (Fig. 2), iv) calculation of variable importance of projection (VIP) values, v) determination of a statistical family by VIP value criterion, vi) significance tests with the false discovery rate (FDR) correction method, and vii) box‐and‐whisker plotting only for significantly changed metabolites. In the study, PiTMaP was applied to 1) an acetaminophen (APAP)‐induced acute liver injury model and control mice and 2) human meningioma samples with different grades (G1–G3), whose results have demonstrated its feasibility. PiTMaP is thus expected to be the next‐generation universal platform to perform rapid metabolic profiling of biological samples.
Fig. 2. PLS‐DA score and loading plots for the control and APAP‐induced liver injury model mice with a VIP criterion of 1.0. Red: APAP model mice and blue green: control mice in PLS‐DA score plots.
Publication details
Zaitsu K, Eguchi S, Ohara T, Kondo K, Ishii A, Tsuchihashi H, Kawamata T, Iguchi A. PiTMaP: A new analytical platform for high-throughput direct metabolome analysis by probe electrospray ionization/tandem mass spectrometry using an R software-based data pipeline. Analytical Chemistry 92:8514–8522, 2020.